Cross-taxa transferability of Sequence Tagged Microsatellite Site (STMS) primers from Pulses to Peanut (Arachis hypogaea L.)
Department of Agronomy and Horticulture, New Mexico State University, Agricultural Science Center at Clovis, Star Route Box 77, Clovis – NM 88101, USA. Email npuppala@nmsu.edu
Cultivated peanut or groundnut (Arachis hypogaea L.) is an important source for oil and protein. The extensive polymorphism of microsatellite markers makes them an ideal choice for studies in population genetics, diversity analysis, and linkage mapping and marker development. The widespread application of microsatellite markers is limited by the requirement for species-specific primers. Development of novel microsatellites remains a costly and lengthy process despite continuous improvements in its efficiency. Transfer of primers across genera (cross-taxa application) offers an alternative to de novo development in plants, with transfer rates ranging from 35-90% and potential polymorphism rates between 58 and 78%. There are few studies that have used cross-transferred microsatellite loci to address questions related to plant populations. In the present study we explore a large number of microsatellites available in common bean to examine their transferability and validity. The results reveal that cross-taxa microsatellite primers amplified PCR products in peanut, and that some of the amplified bands contained microsatellite repeats in them.
To identify and transfer cross-species primers from common bean to peanut and to confirm the presence of microsatellite repeats in the amplified cross-species polymorphism.
Arachis hypogaea, Cross Species, Groundnut, Microsatellite, Peanut, STMS.
Introduction
The application of molecular markers has proven to be useful in most studies of crop evolution and crop improvement. Plant genetic diversity and conservation can be effectively described, managed and preserved using markers. Molecular diagnostic tools are being developed and applied to gene banks and plant breeding programs, particularly for the identification of accessions, the detection of genetic relationships within and among accessions, the measurement of the structure and quantity of genetic variation, and for the identification and localization of particular genes or DNA sequences. Simple sequence repeats (SSR) loci or microsatellites are arrays of short (1 - 6 bp) tandem DNA repeats found in abundance in plant genomes. Many laboratories have the resources and expertise to conduct SSR marker assays, but developing new markers is cumbersome and also time and resource consuming. Sequence data of various crops have revealed conservation among genomes in the regions flanking the SSR loci. Thus, primers designed based on one crop could be used to detect SSR in related crops. Such homology in the flanking regions of the SSR loci has extended the utility of these markers in related species and genera where no information on SSR is available.
Material and Methods
Plant materials
Peanut plant material consisted of 8 genotypes representing 6 sub species of Arachis hypogaea. The details of the accessions are presented in Table 1. Genomic DNA was extracted from 21-day old seedling, with leaves pooled from 3 - 5 seedlings. Primers were synthesized based on information from the public database for common bean (Yu et al. 1999, 2000). These primers represented various functional genes. PCR amplification was performed in a thermocycler (PE 9700), each 10-Ál reaction mix consisting of 1X PCR buffer, 0.4 mM dNTPs, 2.5 mM MgCl2, 0.10 U Taq Polymerase (Ampli Taq Gold), 2 pmol of each primer, and 10 ng of template genomic DNA. The PCR profile consisted of an initial hot start at 95o C for 12 min; 45 cycles of 94o C for 15 sec, 55oC for 30 sec, and 72oC for 1 min, and a terminal extension step at 72o C for 7 min. Amplified PCR products were run on 2% agarose gels to detect amplification and for further analysis (Fig. 1).
Table 1. Accessions used for detection of microsatellite polymorphism by cross-species amplification.
S. No. |
Accession number |
Origin |
Arachis hypogaea var. aequatoriana | ||
1 |
PI 497615 |
Equador |
2 |
PI 497632 |
Equador |
Arachis hypogaea var. fatigiata | ||
3 |
PI 475914 |
Bolivia |
Arachis hypogaea var. hirsuta | ||
4 |
PI 501296 |
Peru |
Arachis hypogaea var.hypogaea | ||
5 |
PI 468191 |
Argentina |
Arachis hypogaea var. peruviana | ||
6 |
PI 502045 |
Peru |
7 |
PI 502096 |
Peru |
Arachis hypogaea var. vulgaris | ||
8 |
PI 494009 |
|
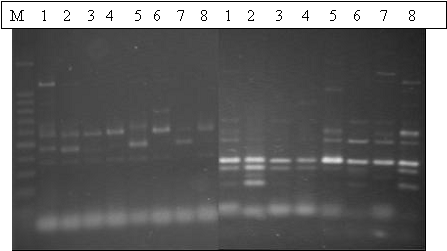
Figure 1. Gel images of two cross specific SSR markers revealing polymorphism in six botanical varieties of peanut.
The polymorphic bands amplified by cross-taxa primers were excised using a clean blade, and the DNA was eluted from the gel using the Qiagen gel extraction kit. The eluted DNA was used as template for reamplification of the putative microsatellite region using corresponding sets of primers and PCR conditions. The re-amplified products were confirmed for amplification and for presence of single band on 2 % agarose gels. The eluted re-amplified DNA was purified using the Qiagen PCR purification kit and used as template for sequencing reaction. The sequencing reaction was conducted using Big Dye Terminator Kit V2.0 (Applied Biosystems). The sequencing reaction products were purified using a sequencing cleanup kit (Edge Biosystems) and the cleaned products were run on an ABI 3100 capillary sequencer.
Results
Out of the 30 cross-taxa SSR primers only 26 primers yielded a PCR product in peanuts. All the successful primers produced multiple bands of high molecular weight in the six sub species of peanut. Polymorphism was detected in many of the PCR products of cross-taxa primers. Re-amplification of the amplified products revealed single to few bands, indicating the presence of internal amplification of the single sequence. Sequence analysis revealed that about 20-25% of the sequences contained microsatellite repeats in the amplified products.
Conclusion
In conclusion microsatellites have become a ‘must’ for many of the genetic studies, but issues related to their isolation are still open. From this study it is evident that cross-taxa amplification could help to isolate repeats from related species, from which more specific repeat-based primers can be designed for further use. The study also revealed that during the evolutionary time scale the repeat type has evolved differently in different genera. A word of caution can be forwarded for future workers regarding the transferability of primers: the amplification of target repeats may be eluding / misleading without prior information about the sequence information within the repeat. The size variation in amplified products may have arisen due to the duplication of the primer-binding site in the target species, thus even leading to the multiple band formation. Since most of the source primers were derived from gene bank of known genes from common bean, it would be interesting to know whether these SSRs have linkage with common bean-related genes.
Acknowledgments
This research was funded in part by grants from the USAID Grant # LAG-G-00-96-90013-00 to the Peanut Collaborative Research Support Program through University of Georgia. Thanks to Mahyco Research Foundation – Hyderabad, India for providing sabbatical leave to the senior author.
References
Yu K, Park SJ and Poysa V (1999) Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 42: 27-34
Yu K, Park SJ, Poysa V and Gepts P (2000). Integration of Simple Sequence Repeats (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). Journal of Heredity 91: 429-434.
Search Site
- Crop Science
- Help/Feedback
- Contact us
- Sitemap





